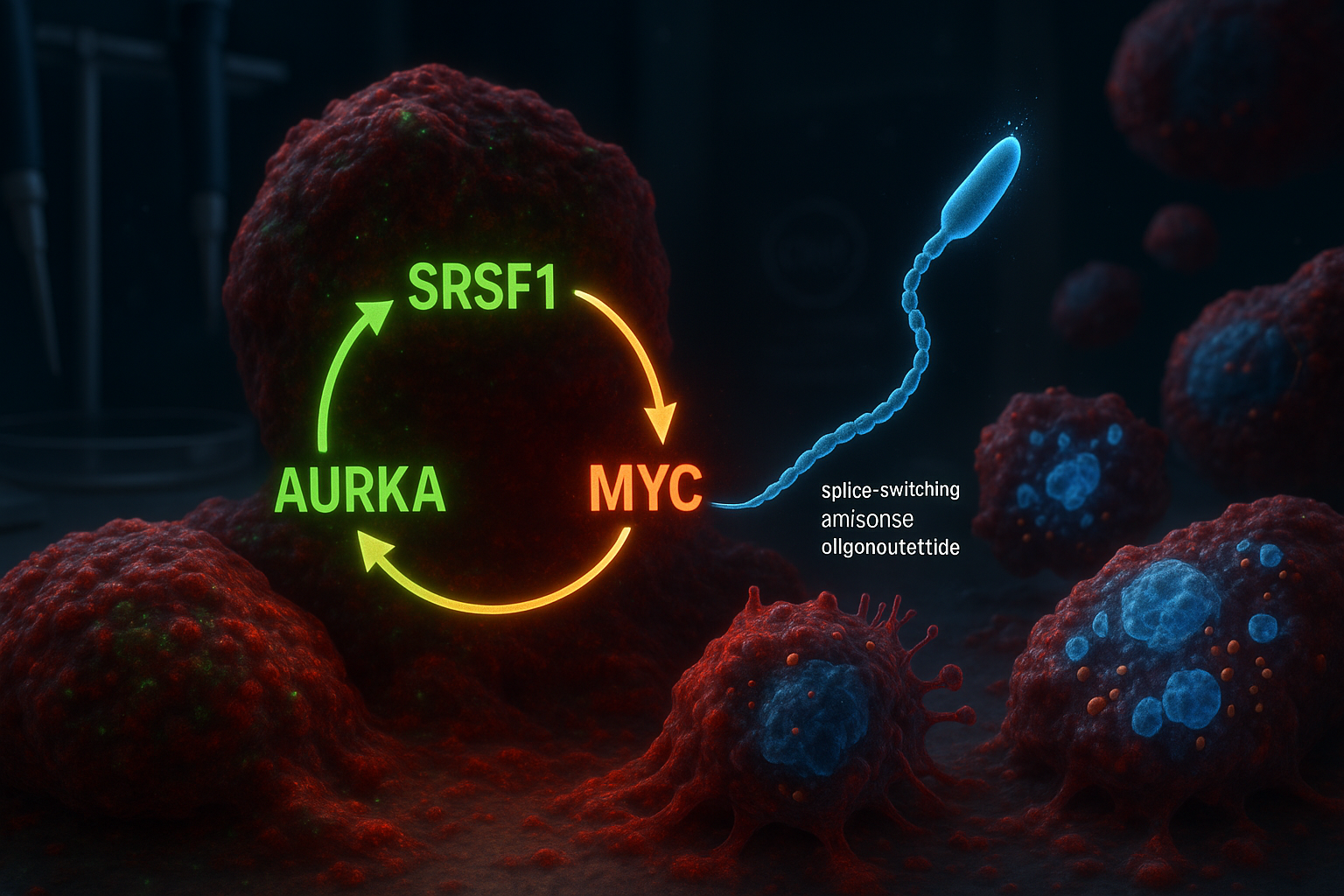
Researchers at Cold Spring Harbor Laboratory report they have identified a three-part molecular circuit involving SRSF1, Aurora kinase A (AURKA) and MYC that helps drive aggressive pancreatic ductal adenocarcinoma. In laboratory models, a splice-switching antisense oligonucleotide designed to alter AURKA splicing disrupted the circuit, reducing tumor-cell viability and triggering programmed cell death.
Pancreatic ductal adenocarcinoma (PDAC) is the most common and deadliest form of pancreatic cancer, and it is notoriously difficult to treat. Many therapeutic strategies have focused on KRAS, a frequently mutated gene in PDAC, but tumors can evade such approaches over time, fueling interest in additional molecular targets.
Cold Spring Harbor Laboratory (CSHL) researchers say their new work builds on earlier findings from the Krainer lab reported in 2023, which identified the RNA-splicing regulator SRSF1 as an early trigger of PDAC tumor formation. By reanalyzing data from that prior study, the team—led by former CSHL graduate student Alexander Kral—concluded that SRSF1 operates in a self-reinforcing circuit with Aurora kinase A (AURKA) and the oncogene MYC.
Within the proposed loop, SRSF1 regulates AURKA through alternative splicing, increasing AURKA levels. AURKA, in turn, helps stabilize the MYC protein, and MYC then boosts production of SRSF1—restarting the cycle and helping drive more aggressive disease behavior.
“Our theory was that some of the changes caused by increased levels of SRSF1 were playing a role in the accelerated tumor growth we were seeing,” Kral said, describing how the team focused on AURKA and then mapped the broader circuit involving MYC.
To try to break the loop, the researchers designed splice-switching antisense oligonucleotides (ASOs) intended to alter how AURKA is spliced. The Krainer lab has long worked on ASO technology and previously helped develop nusinersen (Spinraza), an FDA-approved treatment for spinal muscular atrophy.
In PDAC cell models, the team reported that targeting AURKA splicing produced broader effects than expected: disrupting the circuit was associated with reduced cell viability and the activation of apoptosis.
“It’s like killing three birds with one stone,” senior author Adrian Krainer said. “SRSF1, AURKA, and MYC are all oncogenes contributing to PDAC progression. Just by targeting AURKA splicing with our ASO, we see the loss of these other two molecules as well.”
The study, published in Molecular Cell (online December 30, 2025; listed in the journal’s 2026 volume), is early-stage research and not a clinical treatment. The researchers said further work will be needed to refine the ASO approach before any potential testing in patients.